EpiCypher新推出的DYKDDDDK抗体����,适用于利用FLAG®*标记的靶蛋白进行的研究���。该抗体符合EpiCypher的“CUTANA Compatible”标准���,可用于研究蛋白质-DNA互作的新技术����,CUT & RUN和CUT & Tag�����。每个批次的CUTANA Compatible抗体均使用经EpiCypher优化的方案(https://www.epicypher.com/resources/protocols/)���,按照指定的方法进行测试����,并确定产生的峰值所显示的基因组分布模式与已报道的靶标功能一致��。在过表达 3xFLAG-GATA3的乳腺癌细胞中���,该DYKDDDDK Tag 抗体产生与 GATA3 抗体高度重叠的 CUT&RUN 峰(图 2-3)[1]**��。
*FLAG® is a registered trademark of Merck KGaA, Darmstadt, Germany.
**Thanks to Dr. Takaku (UND) for 3xFLAG-GATA3-3xHA MDA-MB-231 cells.
产品详情
产品名称�����:DYKDDDDK Tag CUTANA™ CUT&RUN Antibody
宿主来源��:Rabbit
实验应用����:CUT&RUN, ELISA, WB, IP, ICC
免疫原����:A synthetic peptide sequence (DYKDDDDK)
克隆性���:Recombinant Monoclonal
保存条件���:Stable for 1 year at 4°C from date of receipt
验证数据
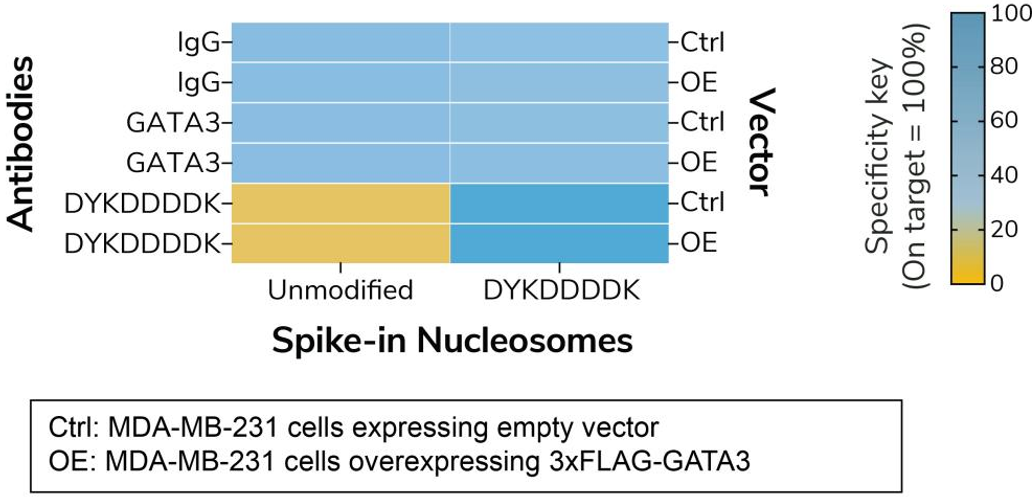
Figure 1: Defined nucleosome spike-ins provide an in-assay control for DYKDDDDK Tag antibody in CUT&RUN |
CUT&RUN was preformed as described in Figure 7. CUT&RUN sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the SNAP-CUTANA™ DYKDDDDK Tag Panel (EpiCypher 19-5001). Data are expressed as a percent relative to on-target recovery (DYKDDDDK Tag set to 100%) or total counts (IgG/GATA3). IgG/GATA3 show no preferential binding to unmodified or DYKDDDDK spike-in nucleosomes. DYKDDDDK Tag antibody selectively enriches the DYKDDDDK Tag spike-in nucleosome, validating the antibody in CUT&RUN.
|
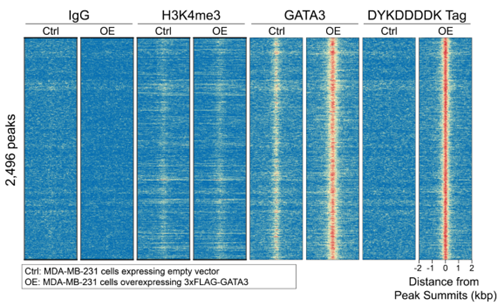
Figure 2: DYKDDDDK-tagged protein peaks in CUT&RUN |
CUT&RUN was preformed as described in Figure 7. Heatmaps show DYKDDDDK Tag antibody-enriched peaks called for FLAG-GATA3 overexpressing cells (OE) in aligned rows relative to all other experimental conditions. Rows are ranked by intensity (top to bottom) and colored such that red indicates high localized enrichment and blue denotes background signal. A high degree of overlap is observed for DYKDDDDK and GATA3 antibodies as expected in the OE cells, while empty vector control (Ctrl) shows absence of FLAG and lower GATA3 enrichment representing endogenous protein. IgG shows low background signal. H3K4me3, a canonical mark of promoters, does not appear in regions of high GATA3 enrichment.
|

Figure 3: CUT&RUN representative browser tracks for DYKDDDDK-tagged protein |
CUT&RUN was preformed as described in Figure 7. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). Two of the top called peaks in FLAG-GATA3 overexpressing (OE) cells are shown. The peaks show the same distribution patterns as observed in the genome-wide heatmaps (Figure 2). IgG shows low background, H3K4me3 is unchanged between empty vector control (Ctrl) and OE cells, GATA3 peaks are more robust in OE cells, and peaks overlap between GATA3 and DYKDDDDK antibodies in OE cells. These results demonstrate the robustness and specificity of DYKDDDDK Tag antibody in CUT&RUN experiments targeting FLAG-tagged proteins.
|
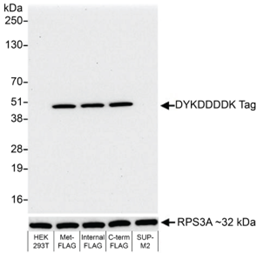
Figure 4: Western blot data |
Western analysis of FLAG-tagged protein in lysates from non-transfected human HEK293 cells, HEK293 transfected with Met-FLAG-IDO1, HEK293 transfected with IDO1-FLAG-HIS (Internal Tag), HEK293 transfected with IDO1-HIS-FLAG (C-Terminal Tag), and SUP-M2 cells. EpiCypher DYKDDDDK Tag antibody was used at 1:1,000. An HRP-conjugated goat anti-rabbit IgG antibody (Fortis A120-101P) was used as a secondary antibody. Chemiluminescence exposure time was 10 seconds. Lower panel shows rabbit anti-RPS3A antibody results (Fortis A305-003A).
|
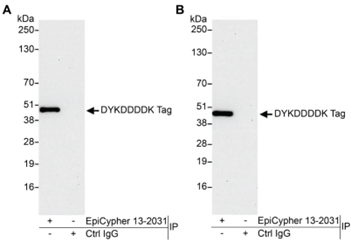
Figure 5: Immunoprecipitation data |
EpiCypher DYKDDDDK Tag antibody (20 µL/mg lysate) was used to immunoprecipitate lysates isolated from HEK293 cells transfected with either Met-FLAG-IDO1 (A) or IDO1-HIS-FLAG (C-terminal tag; B). For blotting immunoprecipitated DYKDDDDK Tag, EpiCypher DYKDDDDK Tag antibody was used at 1:1,000. Chemiluminescence exposure time was 10 seconds.
|
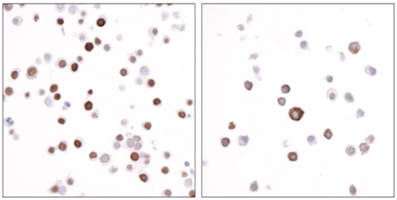
Figure 6: Immunocytochemistry data |
FFPE sections of HEK293 cells expressing either FLAG-tagged nuclear protein (left) or FLAG-tagged cytoplasmic protein (right) using EpiCypher DYKDDDDK Tag antibody. An HRP-conjugated goat anti-rabbit IgG antibody (Fortis A120-501P) was used as a secondary antibody. The substrate used was DAB.
|
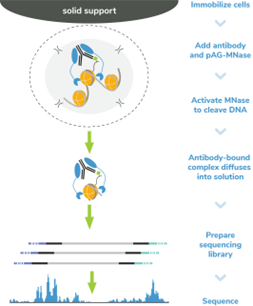
Figure 7: CUT&RUN methods |
CUT&RUN was performed on 500k MDA-MB-231 native cells either stably overexpressing 3xFLAG-tagged GATA3 [1] or containing vector control. Either DYKDDDDK Tag (0.05 µg), H3K4me3 (0.5 µg; EpiCypher 13-0041), GATA3 (0.5 µg; CST 5852), or IgG (0.5 µg, EpiCypher 13-0042) antibodies were used with the CUTANA™ ChIC/CUT&RUN Kit v3 (EpiCypher 14-1048). Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Sample sequencing depth (vector control cells/overexpressing cells) was 5.6/4.5 million reads (IgG), 7.8/6.7 million reads (H3K4me3), 5.8/4.7 million reads (GATA3), and 6.2/5.5 million reads (DYKDDDDK Tag). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
|
订购详情
货号 | 产品名称 | 规格 |
13-2031 | DYKDDDDK Tag CUTANA™ CUT&RUN Antibody | 100 µL |
参考文献
[1] Takaku et al. Genome Biol. (2016). PMID: 26922637

如需了解更多详细信息或相关产品��,请联系EpiCypher中国授权代理商-欣博盛生物
请联系EpiCypher中国授权代理商-欣博盛生物
全国服务热线: 4006-800-892 邮箱: market@qckc0531.com
深圳: 0755-26755892 北京: 010-88594029
广州���:020-87615159 上海: 021-34613729
代理品牌网站: www.qckc0531.com
自主品牌网站: www.neobiosescience.net