EpiCypher新品推荐——H3K4me3 Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag
H3K4me3(组蛋白H3赖氨酸4三甲基化)抗体符合EpiCypher的批次特异性SNAP-Certified™标准���,在CUT&RUN和CUT&Tag应用中具有特异性和高效的靶标富集����。这需要与相关组蛋白 PTMs (使用加标对照的 SNAP-CUTANA™ K-AcylStat Panel 测定�����,EpiCypher 19-1002)的交叉反应性 <20%(图1和图5)����。在不同的细胞起始条件下���,一致的基因组富集结果证实了高靶标效率���:CUT&RUN中500k和50k的细胞量(图2-3)����,CUT&Tag中100k和10k的细胞量(图6-7)�����。即使在细胞数量减少的情况下����,高效抗体也显示出相似的峰结构(图3和7)和高度保守的全基因组信号(图2和6)����。该抗体靶向组蛋白H3K4me3����,其在转录起始位点(TSS)附近的活性启动子处富集并促进基因激活����。
产品详情
产品名称�����:H3K4me3 Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag
宿主来源����:Rabbit
实验应用���:CUT&RUN, CUT&Tag
免疫原��:A synthetic peptide corresponding to histone H3 trimethylated at lysine 4
克隆性��:Monoclonal[2909-3D7]
保存温度����:自收到之日起�����,4℃下可稳定储存1年����。
验证数据
—— CUT&RUN
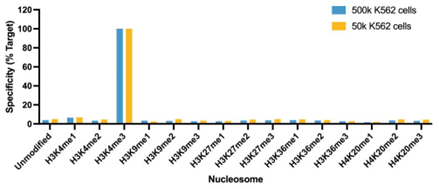
Figure 1: SNAP specificity analysis in CUT&RUN |
| CUT&RUN was performed as described in Figure 4. CUT&RUN sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the K-MetStat panel (x-axis). Data are expressed as a percent relative to on-target recovery (H3K4me3 set to 100%). The antibody showed highly specific recovery of H3K4me3 spike-in nucleosomes at both 500k and 50k cells. |
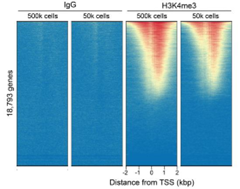
Figure 2: CUT&RUN genome-wide enrichment |
CUT&RUN was performed as described in Figure 4. Sequence reads were aligned to 18,793 annotated transcription start sites (TSSs ± 2 kbp). Signal enrichment was sorted from highest to lowest (top to bottom) relative to the H3K4me3 - 500k cells reaction (all gene rows aligned). High, medium, and low intensity are shown in red, yellow, and blue, respectively. H3K4me3 antibodies produced the expected enrichment pattern, which was consistent between 500k and 50k cells and greater than the IgG negative control.
|
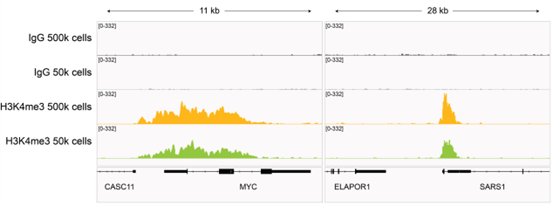
Figure 3: H3K4me3 CUT&RUN representative browser tracks |
| CUT&RUN was performed as described in Figure 4. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). H3K4me3 antibody tracks display sharp peaks at gene promoters, consistent with the biological function of this PTM. Similar results in peak structure and locations were observed for both 500k and 50k cell inputs. |
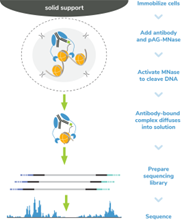
Figure 4: CUT&RUN methods |
| CUT&RUN was performed on 500k and 50k K562 cells with the SNAP-CUTANA™ K-MetStat Panel (EpiCypher 19-1002) spiked-in prior to the addition of 0.5 µg of either IgG negative control (EpiCypher 13-0042) or H3K4me3 antibodies. The experiment was performed using the CUTANA™ ChIC/CUT&RUN Kit v3 (EpiCypher 14-1048). Library preparation was performed with 5 ng of CUT&RUN enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Both kit protocols were adapted for high throughput Tecan liquid handling. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Sample sequencing depth was 3.5 million reads (IgG 500k cell input), 4.0 million reads (IgG 50k cell input), 5.6 million reads (H3K4me3 500k cell input), and 2.8 million reads (H3K4me3 50k cell input). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions. |
—— CUT&Tag
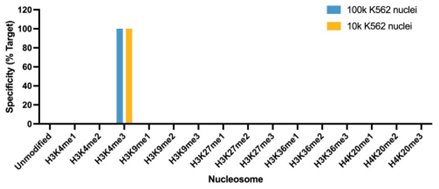
Figure 5: SNAP specificity analysis in CUT&Tag |
| CUT&Tag was performed as described in Figure 8. CUT&Tag sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the K-MetStat panel (x-axis). Data are expressed as a percent relative to on-target recovery (H3K4me3 set to 100%). The antibody showed highly specific recovery of H3K4me3 spike-in nucleosomes at both 100k and 10k nuclei. |
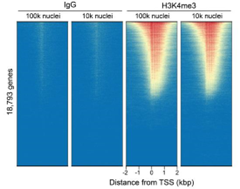
Figure 6: CUT&Tag genome-wide enrichment |
| CUT&Tag was performed as described in Figure 8. Sequence reads were aligned to 18,793 annotated transcription start sites (TSSs ± 2 kbp). Signal enrichment was sorted from highest to lowest (top to bottom) relative to the H3K4me3 - 100k nuclei reaction (all gene rows aligned). High, medium, and low intensity are shown in red, yellow, and blue, respectively. H3K4me3 antibodies produced the expected enrichment pattern, which was consistent between 100k and 10k nuclei and greater than the IgG negative control. |
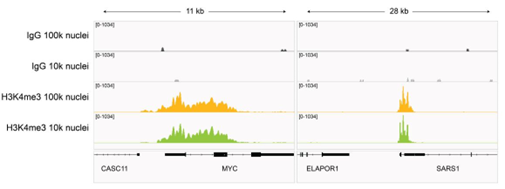
Figure 7: H3K4me3 CUT&Tag representative browser tracks |
| CUT&Tag was performed as described in Figure 8. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). H3K4me3 antibody tracks display sharp peaks at gene promoters, consistent with the biological function of this PTM. Similar results in peak structure and locations were observed for both 100k and 10k nuclei inputs. |
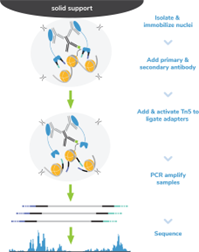
Figure 8: CUT&Tag methods |
CUT&Tag was performed on 100k and 10k K562 nuclei with the SNAP-CUTANA™ K-MetStat Panel (EpiCypher 19-1002) spiked-in prior to the addition of 0.5 µg of either IgG negative control (EpiCypher 13-0042) or H3K4me3 antibodies. The experiment was performed using the CUTANA™ CUT&Tag Kit v1 (EpiCypher 14-1102/14-1103). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Sample sequencing depth was 1.3 million reads (IgG 100k nuclei input), 2.0 million reads (IgG 10k nuclei input), 6.9 million reads (H3K4me3 100k nuclei input), and 10.6 million reads (H3K4me3 10k nuclei input). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
|
订购详情
货号 | 产品名称 | 规格 |
13-0060 | H3K4me3 Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag | 100 µg |

如需了解更多详细信息或相关产品���,
请联系EpiCypher中国授权代理商-欣博盛生物
全国服务热线: 4006-800-892 邮箱: market@qckc0531.com
深圳: 0755-26755892 北京: 010-88594029
广州�����:020-87615159 上海: 021-34613729
代理品牌网站: www.qckc0531.com
自主品牌网站: www.neobiosescience.net